General Services Catalogue
Services commonly provided by MIRRI’s partner organisations, e.g. supply, deposit or identification of microbial resources by gene sequencing, as wll as advanced services, e.g. genomics, screening of metabolites, phylogenetic analysis, consultancy, and up to 95 services. For workflows/pipelines of services, please consult the MIRRI’s Application-specific Service Catalogue.
*All images used are for illustrative purporses only
Deposit of microbiological material that will be publicly available. Used, for example, for new species description
Deposit of microbiological material for patent purposes under the Budapest Treaty
Confidential and private deposit. Access to the microbiological material and associated data has to be authorized by the depositor
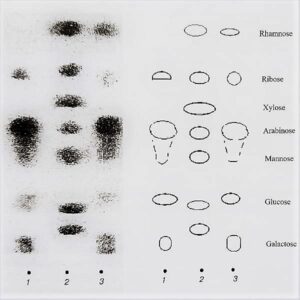
Identification based on the DNA sequence of molecular markers and comparison with curated sequence databases
Identification based on micro- and macroscopic examination combined with the study of different phenotypic and biochemical traits
Identification based on comparison of protein mass spectra generated from unknowns with mass spectra generated from well-characterised reference strains comprised in curated databases
Bioassays on test plants for visual evaluation of symptoms; analysis of morphological and molecular characteristics
Quantitative (qRT-PCR or IFA) or qualitative (PCR) detection and identification of viruses in biological products. Isolation of viruses using cell cultures. Viral genome characterisation (NGS)
The generated STR profile is compared to an appropriate reference sample. If available, DNA from the donor of the tested cell line is the best comparison. If donor DNA can no longer be accessed, the STR profile obtained for the sample is compared to all STR profiles available using an online database.
DNA amplification by PCR of the gene(s) of interest, followed by sequencing of the amplified fragments
DNA amplification by PCR with a single oligonucleotide that hybridizes randomly to the DNA under study, which generates a unique fingerprint of each isolate
DNA amplification by PCR of a fragment from a region of interest (fixed size), followed by separation of the fragments in a gel electrophoresis with an increasing denaturing gradient. This method can detect single base changes/mutations in environmental samples
DNA amplification by PCR of a conserved molecular marker followed by separation of the fragments in a denaturing acrylamide gel electrophoresis under a gradient of temperature. Used to study the microbial diversity of an environmental sample
Digestion of the genomic DNA with specific restriction enzymes followed by amplification by PCR, which generates a unique fingerprint of each isolate
DNA amplification by PCR of microsatellite regions, which generates a unique fingerprint of each isolate
DNA amplification by PCR of extragenic repetitive palindromic elements, which generates a unique fingerprint of each isolate
DNA amplification by PCR of sequences separating Long Terminal Repeats retrotransposons, which generates a unique fingerprint of each isolate
Digestion of the genomic DNA with specific restriction enzymes, which generates a unique fingerprint of each isolate
Digestion of the mitochondrial DNA with specific restriction enzymes, which generates a unique fingerprint of each isolate
DNA amplification by PCR of the 16S rDNA gene, followed by digestion with specific restriction enzymes, which generates a unique fingerprint of each isolate. Can be used to make phylogenetic studies of environmental samples by cloning the 16S rDNA genes of the sample
Digestion of the genomic DNA with specific restriction enzymes and hybridization with a ribosomal (rRNA) probe, to analyse the number and sequence diversity of the rRNA operons
Comparison of protein mass spectra generated from different isolates to cluster them
Quantification of the DNA content in single cells and histogram analysis to estimate the level of ploidy
Detection of fatty acid methyl esters (FAME) by gas chromatography and analysis with specific software (e.g. Sherlock™ Microbial Identification System). Used to differentiate and to characterise isolates
Detection of cell wall polysaccharides using different chromatographic techniques
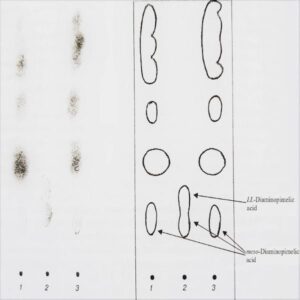
Structural analysis of peptidoglycan by hydrolysis and/or enzymatic digestion of whole-cell extracts coupled with separation of constituents by chromatography and detection by mass spectrometry
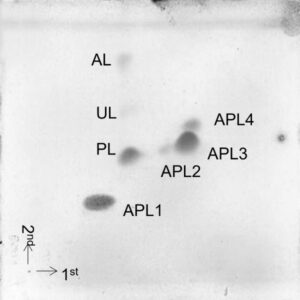
Extraction of the polar lipids (phospholipids, glycolipids, aminolipids) of the cell membrane, followed by chromatographic separation and detection, which provides a pattern of different lipidic compounds
Extraction of the mycolic acids, followed by HPLC separation and mass spectrometry detection. Can be used, for example, to rapidly differentiate Mycobacterium species from Nocardia or Rhodococcus species
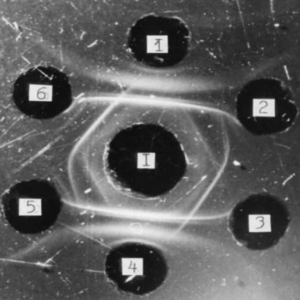
Extraction and purification of the respiratory quinones, followed by HPLC analysis and quantification. Can be coupled with mass spectrometry to analyse complex mixtures. Provides an indication of the type of energy metabolism (aerobic vs anaerobic)
Immunoassays (e.g. ELISA, CL-IA, FIA, LF-IA) used to detect different antigens, including proteins, lipids, sugars, pesticides and other compounds in different types of samples even at low concentrations
High-resolution images of total microbial cells and their ultrathin sections by conventional electron microscopy. Elemental composition of individual cell
Characterisation of the strain based on a variety of assays targeting the metabolic and physiologic profile. The miniaturized systems (e.g. API, Biolog) can be used to tentatively identify the most relevant clinical and industrial microbial species
Cultivation of the strain to induce the production of the enzyme(s), followed by purification/concentration of the intra- or extra-cellular fraction and analysis using substrate-specific probes and/or proteomic approaches
Headspace sampling or metabolite extraction followed by gas chromatography coupled with mass spectrometry analysis/quantification of the volatile fraction
Detection of antioxidant activities on extracts using different reagents
Extraction and purification of the respiratory quinones, followed by HPLC analysis and quantification. Can be coupled with mass spectrometry to analyse complex mixtures. Provides an indication of the type of energy metabolism (aerobic vs anaerobic)
Optimisation of cultures for metabolite production (antimicrobials, pigments, b-glucans, fatty-acids, heterologous proteins expressed in fungi, organic acids, mycotoxins, etc.) and detection/analysis of the metabolite of interest
DNA extraction and next-Generation Sequencing to obtain the raw sequences of the isolate
Quality assessment, reads processing and scaffolding to obtain the draft/complete genome sequence
Quality assessment and reads processing
Quality assessment and reads processing
Strain typing based on the raw sequences or the draft/complete genome sequence (e.g. single-nucleotide polymorphism (SNP) mapping, core genome multilocus sequence typing (cgMLST), G+C content, etc.)
Analysis of the presence of genes to predict phenotypic traits (serotype, antibiotic resistances, virulence factors, production of metabolites, enzymatic activities, etc.)
Pairwise comparison of the genomic sequence of a strain of interest with those of related type strain(s) for identification/classification at the genus/species level
Analysis of the taxonomic composition of an environmental sample or mixed community
Different computational analyses of the genomes on demand, e.g., co-assembly, gene calling, functional taxonomic profiling, recovering metagenome-assembled genomes (MAGs), phylogenomic comparison and profiling, pangenomics
Cultivation in specific media and conditions to separate isolates from mixed samples
Freeze-drying of microbial strains cultures or biological samples for long-term preservation or preparation of samples for further analysis
Customised optimisation of preservation conditions of microbial cultures maximising genetic stability and efficient recovery
Customised optimisation of cultivation and fermentation conditions of microbial cultures to enhance growth rates and productivity
Estimation of the number of cells/virus (total, viable, spores, mycorrhizal infective propagules etc.) present in a culture or sample
Qualitative or quantitative measurements of microbial growth in the presence of compounds or extracts
Test the resistance of a particular strain to different antibiotics
Test the effect of microorganisms, microbial extracts or other agents on the plant fitness infected with a particular pest
Test the effect of microorganisms, microbial extracts, soils or other agents on the plant growth/yield
Test the effect of microorganisms, microbial extracts or other agents on the viral fitness
Metabolomic analysis on multiwell plates using a UV/VIS fluorescence microplate reader
Viability estimation under different conditions measuring the absorbance of cultures stained with iodonitrotetrazolium violet on multiwell plates
Analysis of adhesive activities of strains stained with crystal violet on multiwell plates
Assessment of the productivity, robustness, harvesting characteristics and/or production of metabolites (bioproducts, bioactive substances, enzymes, etc.) of a strain culture
Analysis of the capacity of microbial strains to adhere to different surfaces
Analysis of the capacity of microbial strains to produce molecules with biosurfactant and emulsion activity
Cultivation protocols adapted to the test materials and products are used to evidence contaminating microorganisms
The resistance of diverse materials against microbial damage is analysed using growth tests and control strains
Assessment of the metabolic status of a microbial strain analysing the O2 consumption / CO2 production ratio
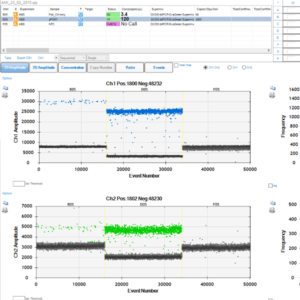
Quantification of absolute count of DNA or RNA using Droplet digital PCR
Phenotypic and genome mining based assessment of key traits recommended by the European Food Safety Agency (EFSA) (e.g., presence of virulence, bacteriocin and antimicrobial resistance genes; presence of elements involved in horizontal gene transfer; antimicrobial resistance assays)
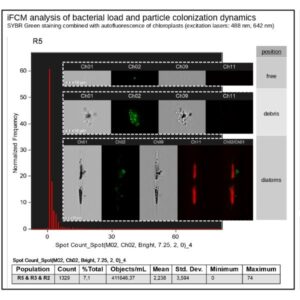
Combination of microscopy and flow cytometry providing bright-field, dark-field (SSC), and several fluorescent images of each cell in flow, at high speed. It can be used for spatial localisation of biomolecules and cellular components, reporting signal intensity with very high sensitivity.
Separation of cell populations based on different features such as size and granularity (via light scattering) or fluorescence characteristics (fluorescence detector). Separated cells remain viable and can be cultured after the process
Genomic DNA extraction from a pure culture of the microorganism suited for subsequent analysis (e.g. PCR, WGS)
Conjugation of cells/spores by micromanipulation and analysis of the resulting zygotes (flow cytometry, electrophoretic karyotyping, marker analysis, stability, etc.)
Variety of training courses related to several topics (isolation, cultivation, preservation, identification, characterisation and valorisation of microbiological material, plasmid DNA handling techniques, quality management and operation of culture collections, etc) and also tailored training courses adapted to the specific needs of the user. See more details here.